Multicellular Dynamics during Development
Hello!~
My name is Guoye Guan (关国业) and I’m a biophysicist and systems biologist. I am studying the embryonic, synthetic, and microbial systems with both computational approaches and experimental data. I completed my Ph.D. on worm development at Prof. Chao Tang's Lab (Peking University) and am a PostDoc. on human development at Prof. Sahand Hormoz Lab (Harvard Medical School & Dana-Farber Cancer Institute).
Research Projects
Exploring developmental patterns at molecular-to-multicellular scales through biophysics and systems biology approaches.
DL/AI-Driven Multidimensional
Developmental Atlas Analysis
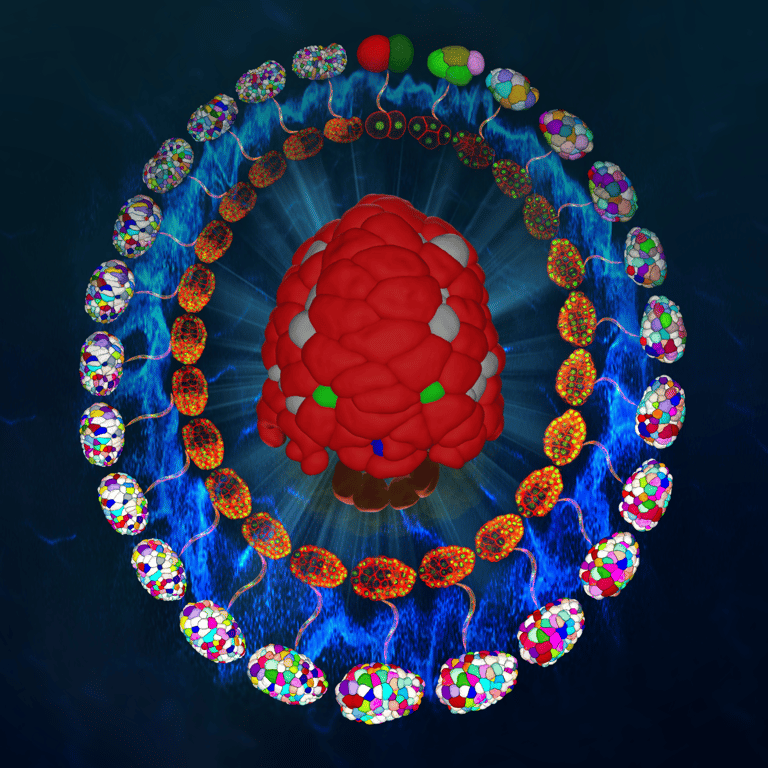
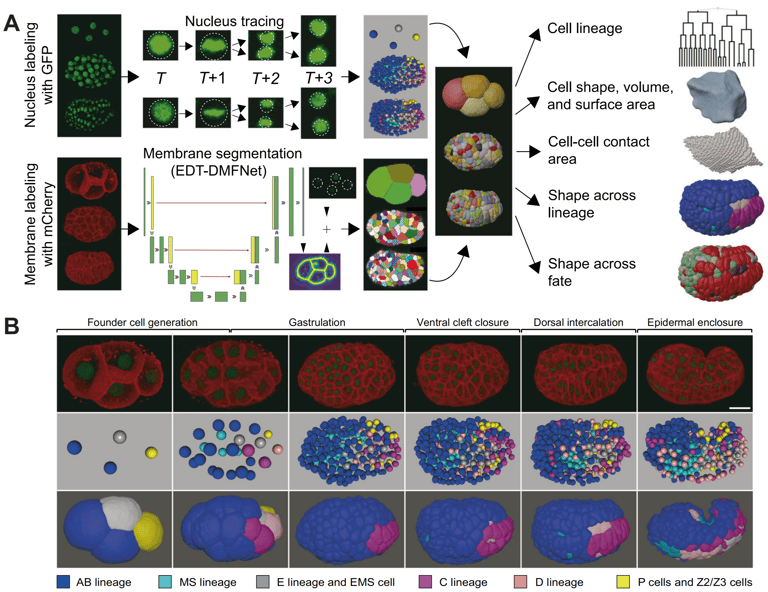
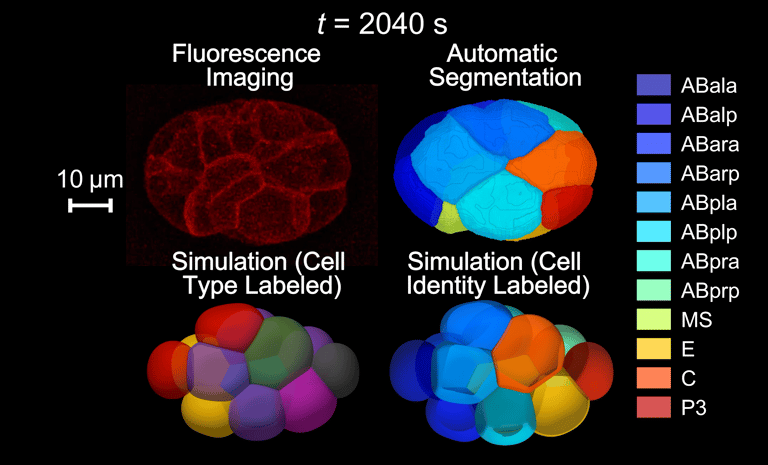
Using human and worm systems, we apply deep learning and artificial intelligence to extract information from development — quantifying data from microscopic images (e.g., molecular accumulation, cell movement, embryo/embryoid morphology) and inferring regulations from developmental dynamics.
Related Works:
Guan G†, Li Z†, Ma Y†, Ye P†, Cao J, Wong MK, Ho VWS, Chan LY, Yan H*, Tang C*, Zhao Z*. Cell lineage-resolved embryonic morphological map reveals novel signaling regulating cell fate and size asymmetry. Nat. Commun. (2025)
Cao J†, Guan G†, Ho VWS†, Wong MK, Chan LY, Tang C*, Zhao Z*, Yan H*. Establishment of a morphological atlas of the Caenorhabditis elegans embryo using deep-learning-based 4D segmentation. Nat. Commun. (2020)
Mammalian-Based Developmental Pattern
Design and Synthesis
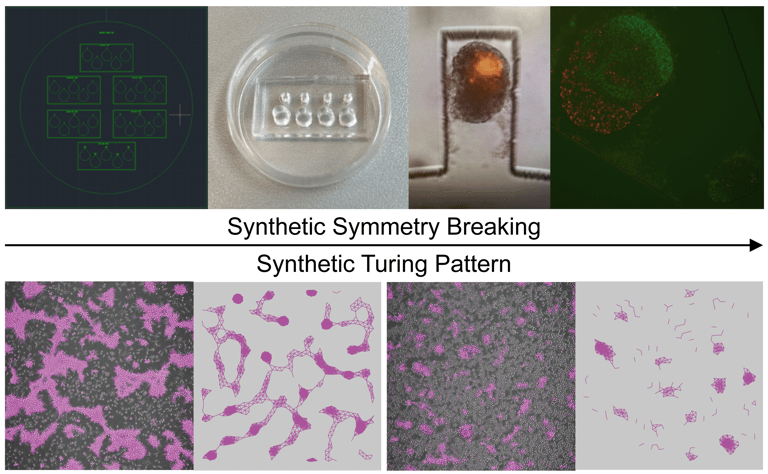
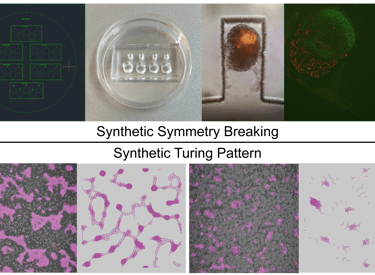
Using human and mouse systems, we design and synthesize developmental patterns —both those found in nature and those that do not occur naturally — such as symmetry breaking, hexagonal lattice, and stripe- or spot-like Turing patterns. The synthetic patterns could be self-organized in well plates or be reinforced within microfluidic chips. We use physical models and virtual experiments to provide rational guidance for synthesizing developmental patterns. Our in vitro and in silico methodology seeks to uncover key processes in natural development while engineering tissues and organs with customized spatial functions, such as hair follicles.

Cell Polarity and Division
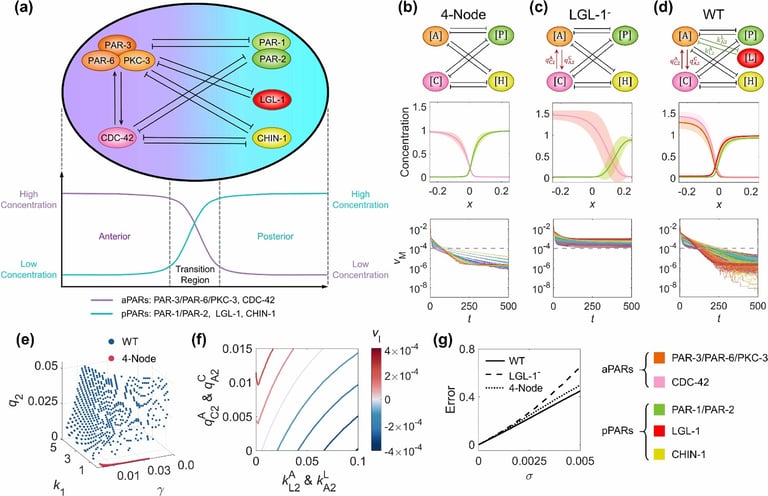
Using physical models, we simulate cell-division-related biological processes across multiple scales, from molecular interaction networks, cell membrane curvature and mechanics, to cell proliferation and differentiation. Through theoretical and numerical analyses, we aim to uncover the design principles underlying these unique functional processes, providing guidance for restoring or constructing desired biological functions, such as developmental robustness and speed.
Related Works:
Chen Y†, Guan G†, Tang LH*, Tang C*. Balancing reaction-diffusion network for cell polarization pattern with stability and asymmetry. eLife (2025)
Guan G†, Wong MK, Zhao Z, Tang LH*, Tang C*. Volume segregation programming in a nematode's early embryogenesis. Phys. Rev. E (2021)
Cell Mechanics and Morphogenesis
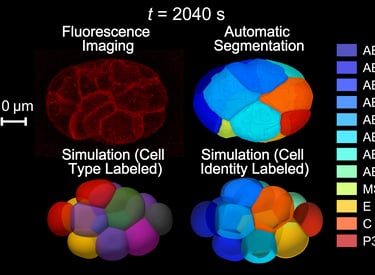
By combining time-lapse 3D microscopic imaging with deep learning/artificial intelligence-based data reconstruction, we develop multi-scale physical models to understand how cells mechanically interact during developmental morphogenesis. This approach enables the creation of a digital life — a digital twin of real life — that can be freely dissected and perturbed on a computer. This allows experiments that are otherwise difficult or impossible in reality. Aligning the behaviors of real and digital life helps identify accurate physical representations of living phenomena at the microscopic scale.
Related Works:
Guan G†, Kuang X†, Tang C*, Zhang L*. Comparison between phase-field model and coarse-grained model for characterizing cell-resolved morphological and mechanical properties in a multicellular system. Commun. Nonlinear Sci. Numer. Simul. (2023)
Kuang X†, Guan G†, Wong MK, Chan LY, Zhao Z, Tang C*, Zhang L*. Computable early Caenorhabditis elegans embryo with a phase field model. PLoS Comput. Biol. (2022)
Image Credit: Guoye Guan, Christoph Budjan, Xuanqi Zhang, Satoshi Toda
Image Credit: Guoye Guan, Zelin Li, Yiming Ma, Pohao Ye
Image Credit: Guoye Guan, Xiangyu Kuang, Qianli Zhang
Image Credit: Guoye Guan, Yixuan Chen
Contact Me
For inquiries about my research, please reach out through the contact information below.
© 2025. All rights reserved.
Guoye Guan (关国业), Ph.D.
Department of Systems Biology, Harvard Medical School
Department of Data Science, Dana-Farber Cancer Institute
Office: 11054/Wet Lab, Center for Life Science Boston, 3 Blackfan St, Boston, MA, USA
Phone: (+1) 617-963-6484
Email: guanguoye@gmail.com/guoye_guan@dfci.harvard.edu/guoye@ds.dfci.harvard.edu